Data Description: Family Relations
Review: we have identified genetic and social pedigrees and the relation of the iGENE Sample members to all of their household members.
Goal: Now we want to look at some descriptives of what we build.
preparation: read data needed in the script
# data of igene-sample
ppfad_igene_long <- import(here::here("data/ppfad_igene_long_v36.rds"))
pid_igene_sample <- import(here::here("data/pid_igene_sample.rds")) %>% tibble(pid = .)
igene_sample <- import(here::here("data/igene_sample.rds"))
soepis_igene_raw <- import(here::here("data/soepis_igene_raw.rds"))
# data with parental pointers
social_pedigree <- import(here::here("data/social_pedigree_v36.rds"))
social_pedigree_justigene <- import(here::here("data/social_pedigree_justigene_v36.rds"))
genetic_pedigree <- import(here::here("data/genetic_pedigree_v36.rds"))
genetic_pedigree_justigene <- import(here::here("data/genetic_pedigree_justigene_v36.rds"))
igene_hhmember_relations <- import(here::here("data/igene_hhmember_relations_v36.rds"))
igene_hhmember_relations_justigene <- import(here::here("data/igene_hhmember_relations_justigene_v36.rds"))describe sample
Number of observations
N ppfad_igene_long = 2602 N ppfad_igene_long = 20575
ppfad_igene_long %>%
count(syear) %>%
mutate(label = if_else(syear %in% c("1998", "2009", "2019"), n, NA_integer_)) %>%
ggplot(aes(syear, n, label = label))+
geom_col()+
labs(title = "Number of iGENE-Sample Members over Time",
x = "Survey Year", y = "") +
geom_label(size = 3, vjust = -0.1)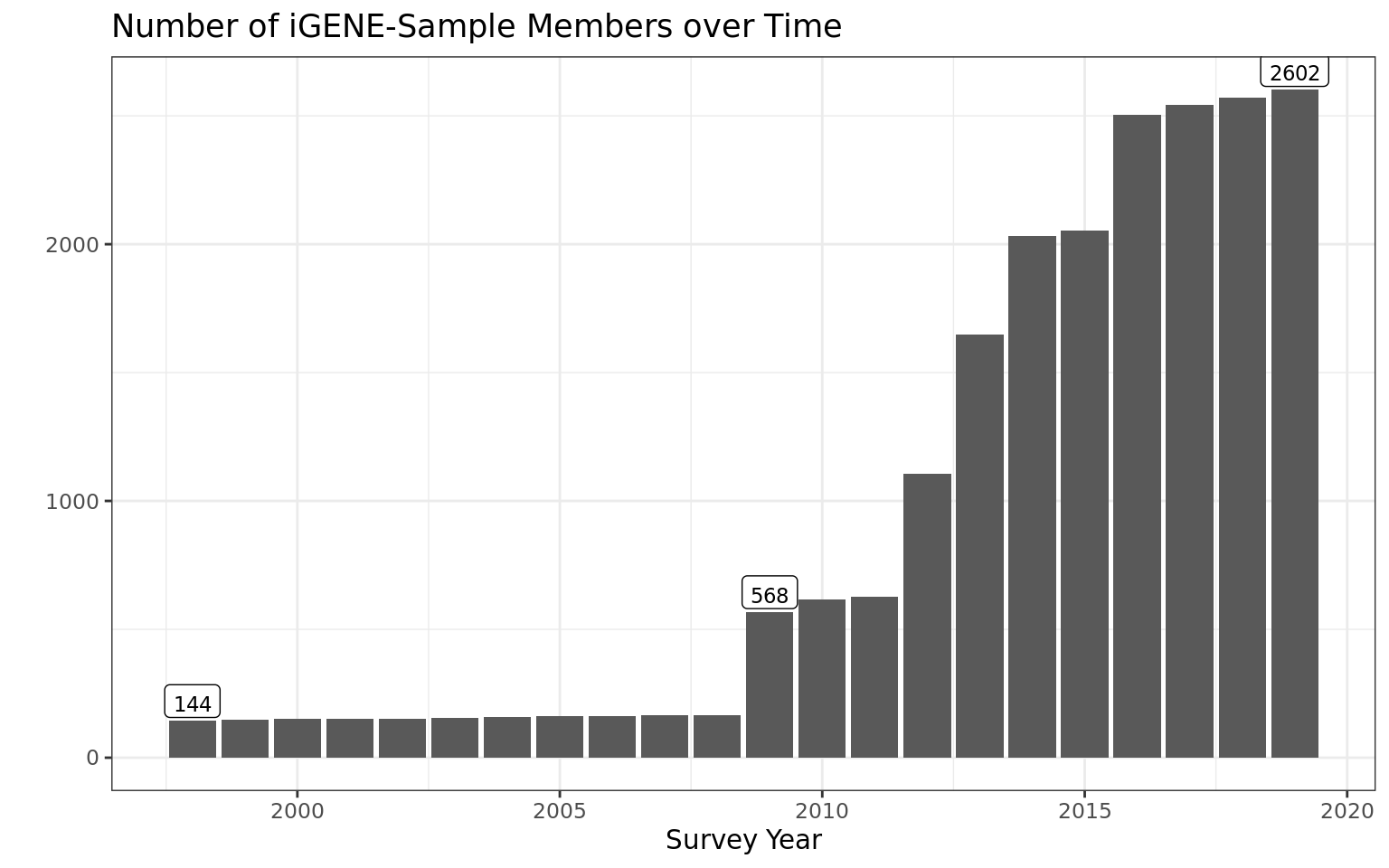
Sample Membership
ppfad_igene_long %>%
arrange(desc(syear)) %>%
distinct(pid, .keep_all = T) %>%
# filter out missing years and individuals
filter(netto > 0) %>%
tabyl(netto_l, show_missing_levels = F) %>% arrange(desc(n)) %>% adorn_pct_formatting()soepis_igene_raw %>%
mutate_all(sjlabelled::as_label) %>%
tabyl(probe, igene15_kind)Relationships to Household Members
igene_hhmember_relations %>%
distinct(pid, .keep_all = T ) %>%
semi_join(pid_igene_sample, by = c("match_id" = "pid")) %>%
drop_na(igene_stell) %>%
tabyl(igene_stell_l, show_missing_levels = FALSE) %>%
arrange(desc(n)) %>%
adorn_pct_formatting() %>% adorn_totalsHow many other relatives live with sample members?
igene_hhmember_relations %>%
# semi_join(pid_igene_sample, by = c("match_id" = "pid")) %>%
filter(!str_detect(igene_match_rel, "missing")) %>%
tabyl(igene_match_rel) %>%
arrange(desc(n)) %>%
adorn_pct_formatting()igene_hhmember_relations %>%
distinct(pid, match_id, .keep_all = T) %>%
filter(pid != match_id) %>%
semi_join(pid_igene_sample, by = c("match_id" = "pid")) %>%
filter(!str_detect(igene_match_rel, "missing")) %>%
tabyl(igene_match_rel, igene_match_type_minor) %>%
adorn_title(row = "iGENE Member is ... of HH-Member",
col = "HH-Member Relation")igene_hhmember_relations %>%
distinct(pid, match_id, .keep_all = T) %>%
filter(pid != match_id) %>%
filter(!str_detect(igene_match_rel, "missing")) %>%
tabyl(igene_match_rel, igene_match_type_major, match_gender) %>%
adorn_title(row = "iGENE Member is ... of HH-Member",
col = "HH-Member Relation")## $female
## HH-Member Relation
## iGENE Member is ... of HH-Member genetic social
## child 489 13
## grandchild 8 8
## grandgrandchild 1 0
## grandparent 14 4
## other 0 50
## parent 629 39
## partner 600 204
## relative 14 3
## sibling 306 20
##
## $male
## HH-Member Relation
## iGENE Member is ... of HH-Member genetic social
## child 412 18
## grandchild 9 2
## grandgrandchild 0 0
## grandparent 11 3
## other 0 38
## parent 766 56
## partner 375 519
## relative 16 0
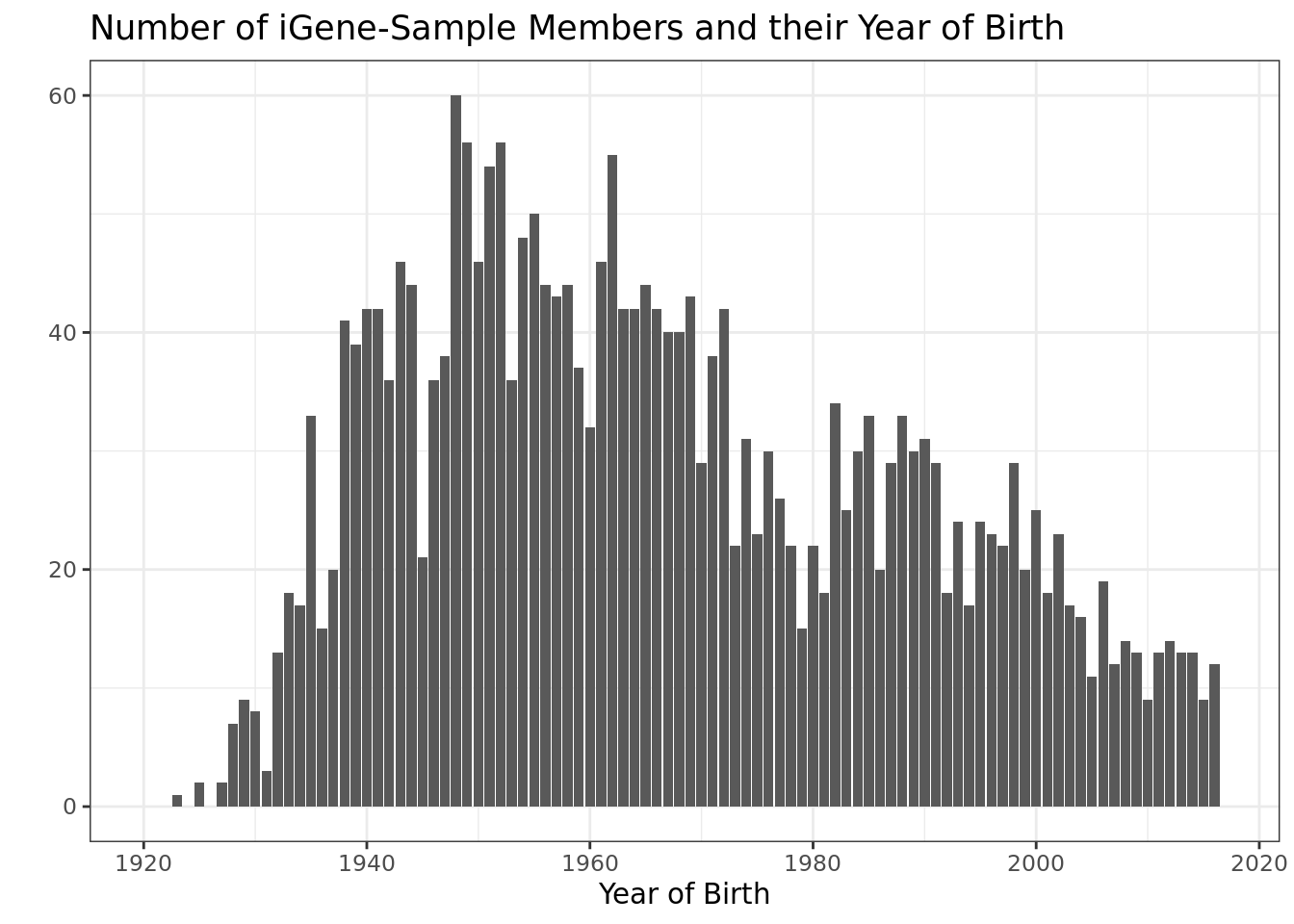
## sibling 282 35Year of Birth
igene_sample %>%
ggplot(aes(lgeb)) +
geom_bar() +
labs(y = "",
x = "Year of Birth",
title = "Number of iGene-Sample Members and their Year of Birth") +
xlim(1920, 2017)