Plot: SOEP-IS Sample
# load data
# soepisv34 <- rio::import(here::here("data/soepisv34.rds"))
soepis_long <- rio::import(here::here("data/soepis_long_v36.rds"))
soepis_age <- rio::import(here::here("data/soepis_age_v36.rds"))Explore Data for Gene Analyis
All variables
By Year
First you can see the number of available observation for each variable in each year
- x-axis = survey year
- y-axis = variables
- size = number of observations
- colour = variable group
soepis_long %>%
drop_na(value) %>%
# filter(row_number() < 1000) %>%
mutate(key_name_label = factor(key_name_label),
order = as.numeric(key_category),
key_name_label = fct_reorder(key_name_label, desc(order))) %>%
ggplot(aes(key_name_label, syear, col = key_category)) +
geom_count() +
coord_flip() +
theme(legend.position="right") +
guides(col = guide_legend(ncol = 1)) +
scale_x_discrete(labels = wrap_format(40))+
scale_y_continuous(limits= c(1998, 2019), breaks = seq(1998,2020,2))+
labs(title = "Number of observations for selected SOEP variables from 1998 - 2019",
subtitle = "Size indicates number of observations",
y = "", x = "")
By Age
soepis_age %>%
drop_na(value) %>%
drop_na(age) %>%
# filter(row_number() < 1000) %>%
mutate(key_name_label = factor(key_name_label),
order = as.numeric(key_category),
key_name_label = fct_reorder(key_name_label, desc(order))) %>%
ggplot(aes(key_name_label, age, col = key_category)) +
geom_count() +
# scale_size(breaks = seq(0, max(n), 10000)) +
coord_flip() +
theme(legend.position="right") +
guides(col = guide_legend(ncol = 1)) +
scale_x_discrete(labels = wrap_format(40))+
scale_y_continuous(limits= c(00, 110), breaks = seq(0,110,10))+
labs(title = "Number of observations for selected SOEP variables by age",
subtitle = "Size indicates number of observations",
y = "", x = "")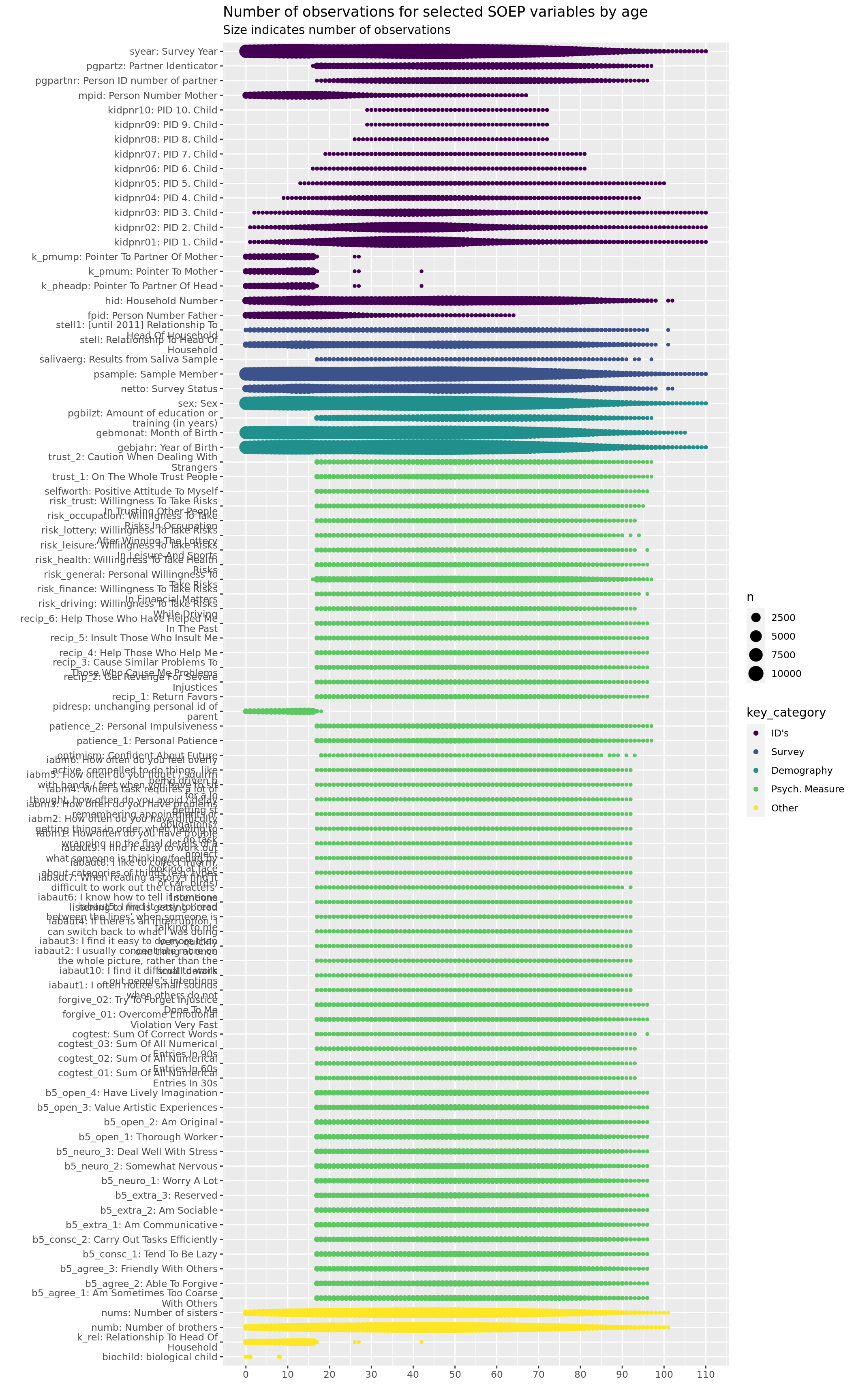
soepis_age %>%
drop_na(value) %>%
# filter(row_number() < 1000) %>%
filter(age >= 0) %>%
mutate(key_name_label = factor(key_name_label),
order = as.numeric(key_category),
key_name_label = fct_reorder(key_name_label, desc(order)),
age_k = cut_width(age, 15, boundary = 0)) %>%
ggplot(aes(key_name_label, age, col = key_category)) +
geom_count() +
# scale_size(breaks = seq(0, max(n), 10000)) +
coord_flip() +
theme(legend.position="right") +
guides(col = guide_legend(ncol = 1)) +
scale_x_discrete(labels = wrap_format(40))+
scale_y_continuous(limits= c(00, 110), breaks = seq(0,110,10))+
labs(title = "Number of observations for selected SOEP variables by age",
subtitle = "Size indicates number of observations",
y = "", x = "") +
facet_wrap(~ age_k, scales = "free_x", nrow = 1, drop = T)+
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
legend.position = "top") +
guides(size = guide_legend(order = 2, nrow = 1),
col = guide_legend(nrow = 1, title = "Category"))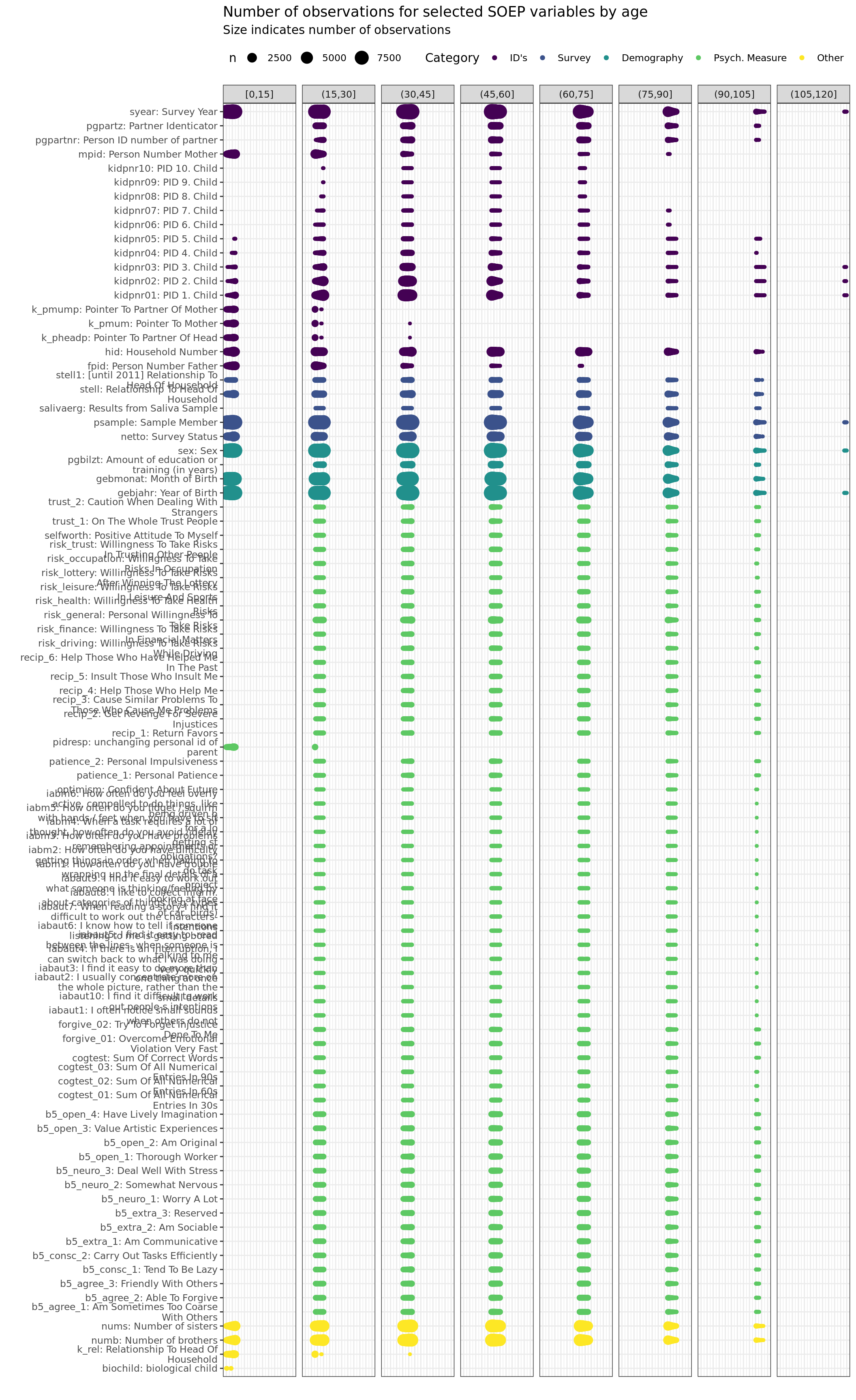
soepis_age %>%
drop_na(value) %>%
# filter(row_number() < 1000) %>%
filter(age >= 0) %>%
mutate(key_name_label = factor(key_name_label),
order = as.numeric(key_category),
key_name_label = fct_reorder(key_name_label, desc(order)),
age_k = cut_width(age, 15, boundary = 0)) %>%
ggplot(aes(key_name_label, age, col = key_category)) +
geom_count() +
# scale_size(breaks = seq(0, max(n), 10000)) +
coord_flip() +
theme(legend.position="right") +
guides(col = guide_legend(ncol = 1)) +
scale_x_discrete(labels = wrap_format(40))+
scale_y_continuous(limits= c(00, 110), breaks = seq(0,110,10))+
labs(title = "Number of observations for selected SOEP variables by age",
subtitle = "Size indicates number of observations",
y = "", x = "") +
facet_wrap(~ age_k, scales = "free_x", nrow = 1, drop = T)+
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank())
Overall
Here is an overall plot of the number of available observations for each of the variables. It helps to get a general understanding of the proportions of missings for groups of variables
soepis_long %>%
drop_na(value) %>%
filter(key_category != "Psych. Measure") %>%
group_by(key) %>%
add_count() %>%
ungroup() %>%
distinct(key, .keep_all = T) %>%
group_by(key_category) %>%
mutate(key_name_label = fct_reorder(factor(key_name_label), n)) %>%
ggplot(aes(x = key_name_label, y = n, fill = key_category, label = n)) +
geom_col(width = 0.2) +
geom_point() +
geom_label(color = "white", size = 2) +
coord_flip() +
scale_y_continuous(labels = scales::label_number_auto()) +
scale_x_discrete(labels = wrap_format(40))+
theme_light() +
theme(legend.position = "none") +
facet_wrap(~key_category, ncol = 1, scales = "free") +
labs(title = "Overall Number of observations for selected SOEP variables from 1998 - 2019", y = "", x = "") 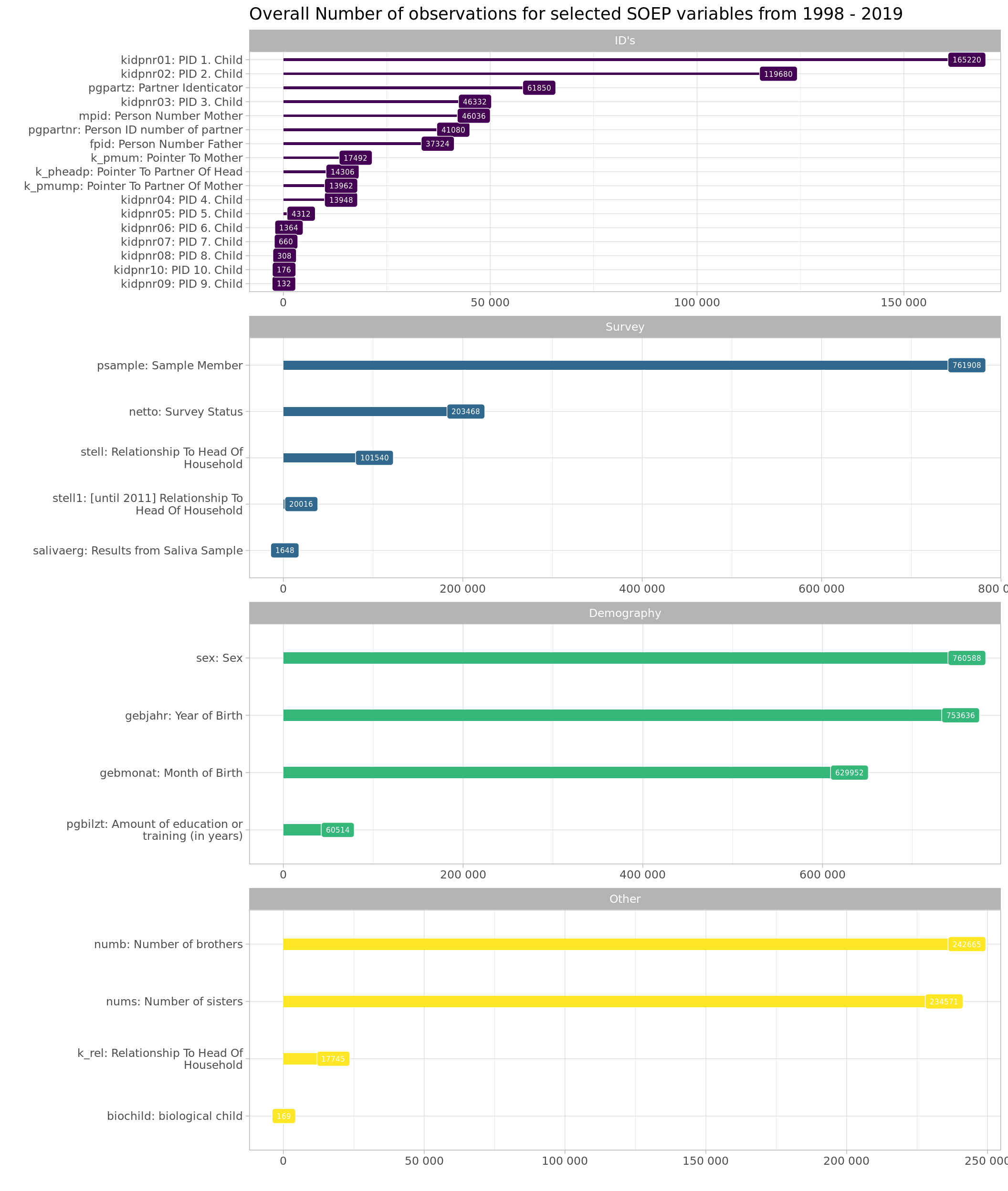
soepis_long %>%
drop_na(value) %>%
filter(key_category == "Psych. Measure") %>%
group_by(key) %>%
add_count() %>%
ungroup() %>%
distinct(key, .keep_all = T) %>%
mutate(key_name_label = fct_reorder(factor(key_name_label), n)) %>%
ggplot(aes(x = key_name_label, y = n, fill = key_category, label = n)) +
geom_col(width = 0.3) +
geom_point() +
geom_label(color = "white", size = 3) +
coord_flip() +
scale_y_continuous(labels = scales::label_number_auto()) +
scale_x_discrete(labels = wrap_format(40))+
theme_light() +
theme(legend.position = "none") +
facet_wrap(~key_category, ncol = 1) +
labs(title = "Overall Number of observations for selected SOEP variables from 1998 - 2019", y = "", x = "")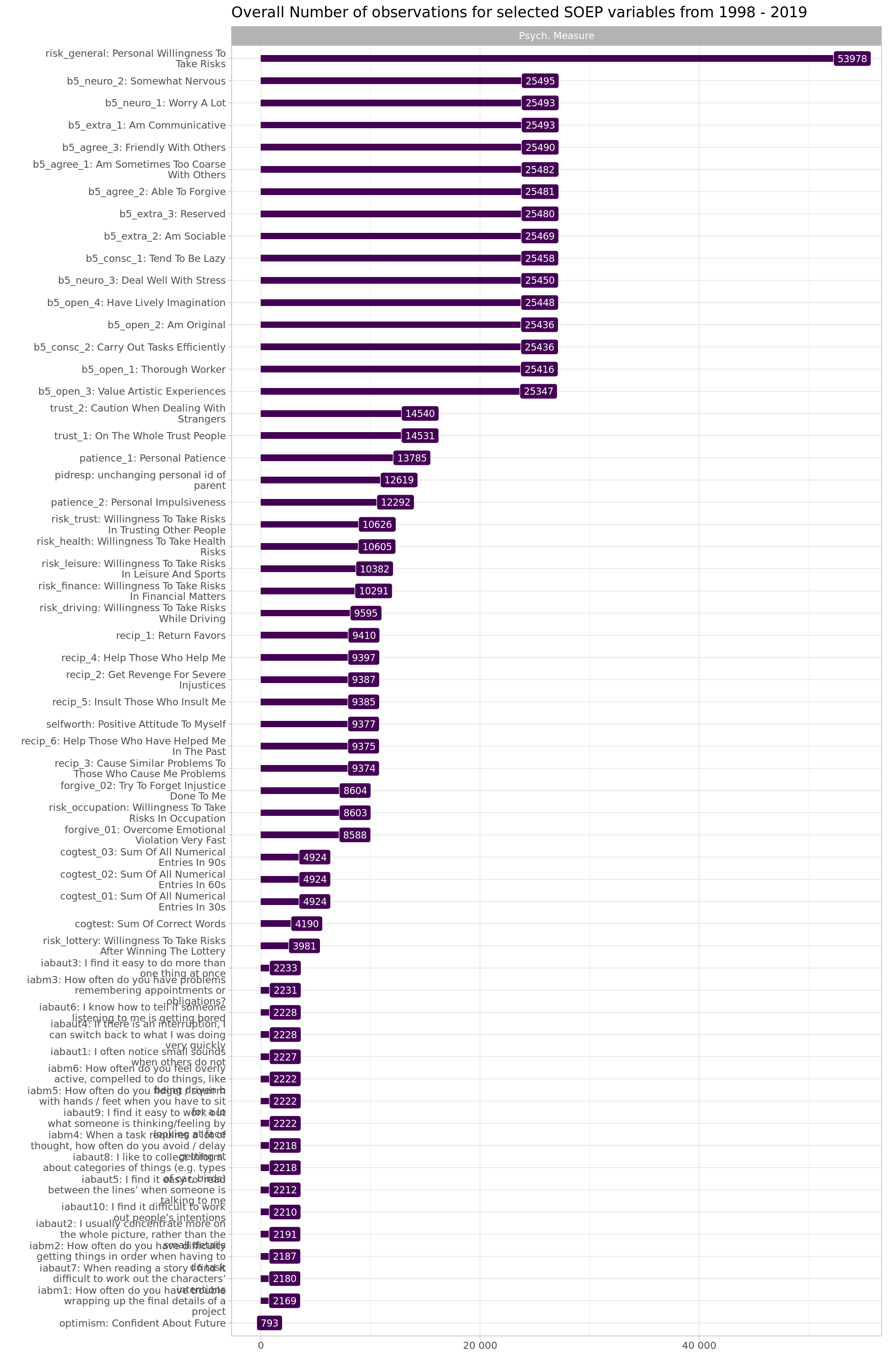
By Variable Category
By Year
ID’s
soepis_long %>%
drop_na(value) %>%
filter(key_category == "ID's") %>%
ggplot(aes(key_name_label, syear)) +
geom_count(col= "#440154FF") +
coord_flip() +
theme(legend.position="right") +
scale_x_discrete(labels = wrap_format(40))+
scale_y_continuous(limits= c(1998, 2019), breaks = seq(1998,2020,2)) +
labs(title = "Number of observations for selected SOEP variables from 1998 - 2019",
subtitle = "Size indicates number of observations",
y = "", x = "") 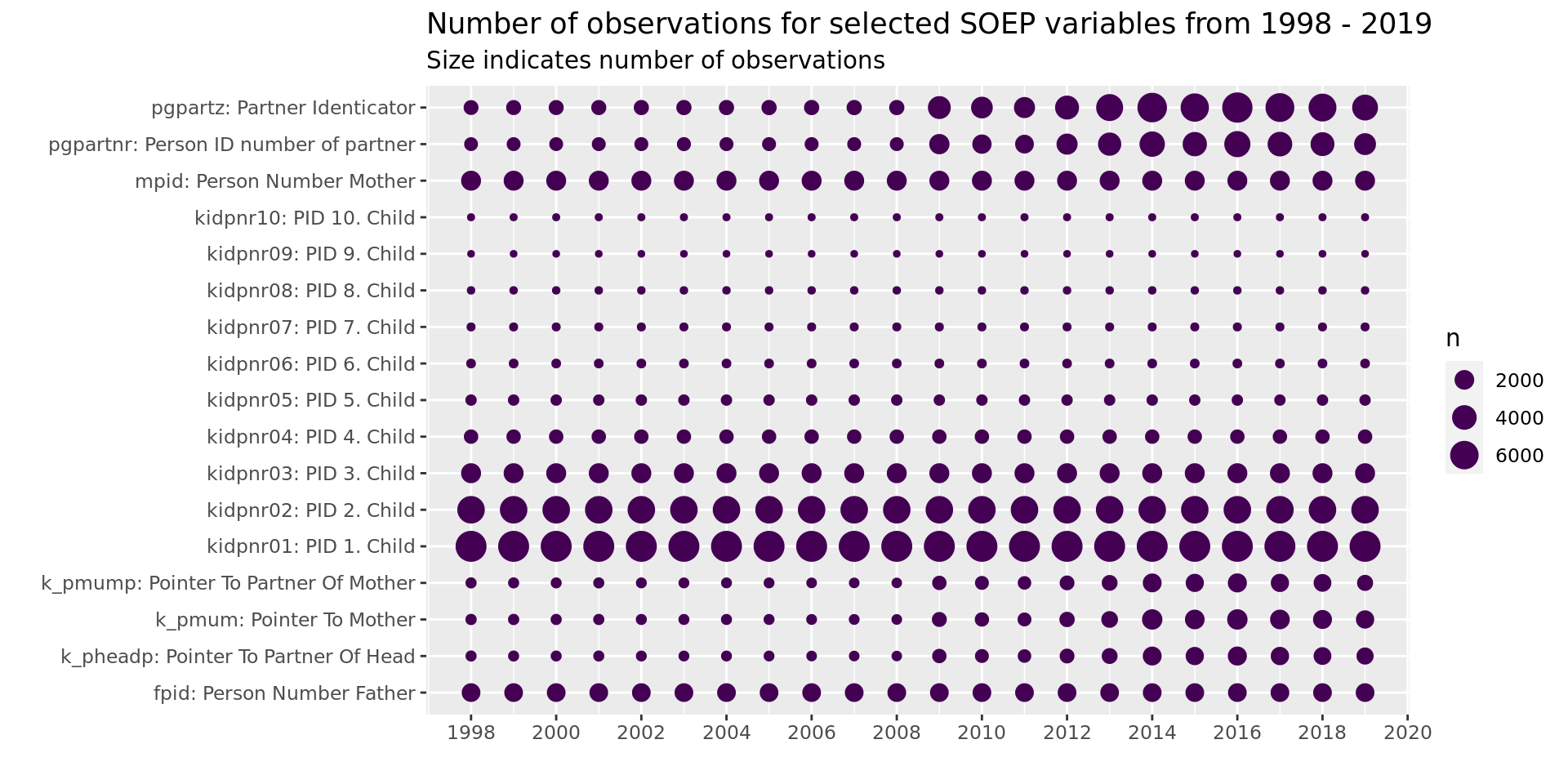
Survey
soepis_long %>%
drop_na(value) %>%
filter(key_category == "Survey") %>%
ggplot(aes(key_name_label, syear)) +
geom_count(col= "#3B528BFF") +
coord_flip() +
theme(legend.position="right") +
scale_x_discrete(labels = wrap_format(40))+
scale_y_continuous(limits= c(1998, 2019), breaks = seq(1998,2020,2)) +
labs(title = "Number of observations for selected SOEP variables from 1998 - 2019",
subtitle = "Size indicates number of observations",
y = "", x = "") 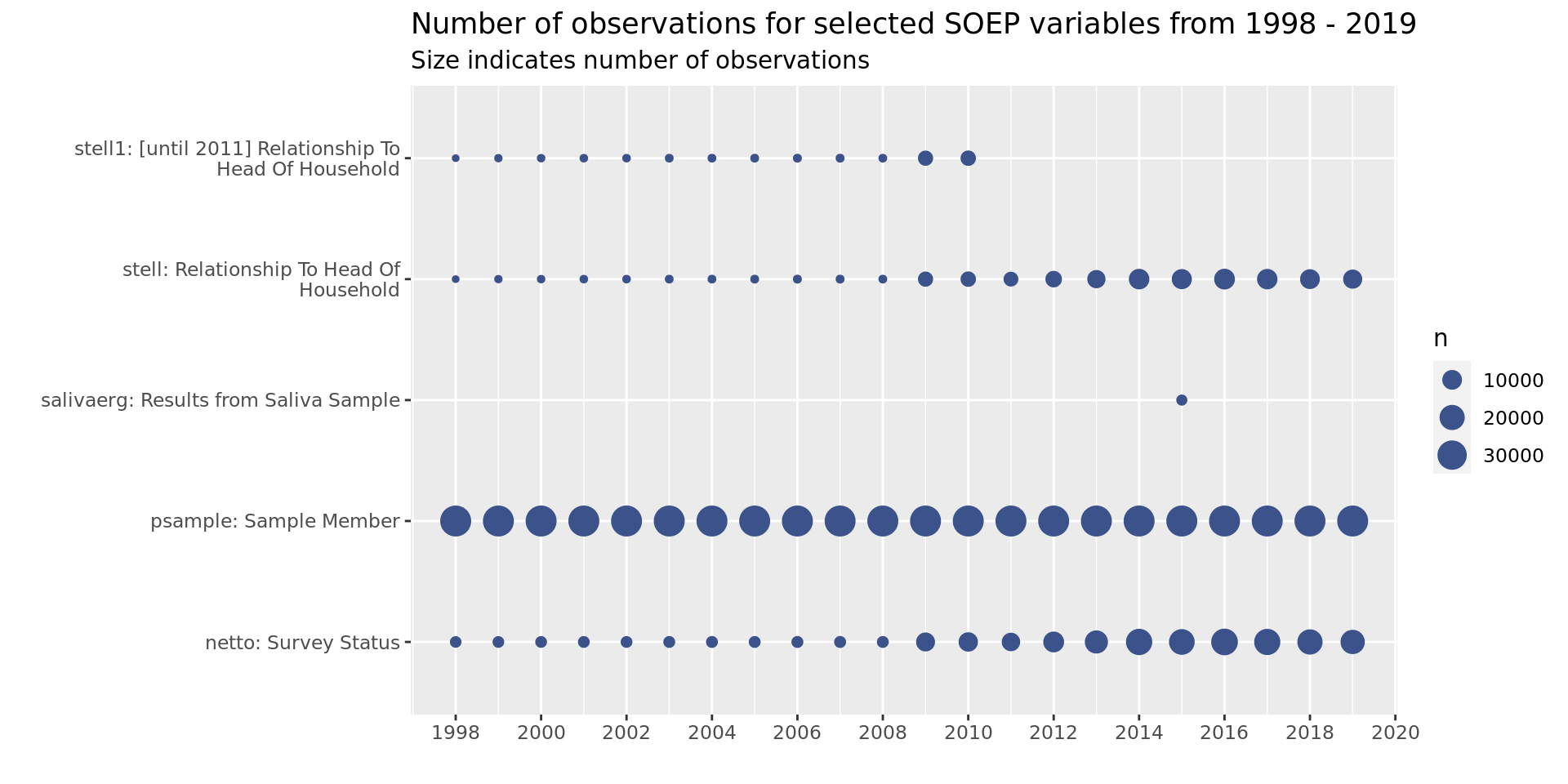
Demography
soepis_long %>%
drop_na(value) %>%
filter(key_category == "Demography") %>%
ggplot(aes(key_name_label, syear)) +
geom_count(col= "#21908CFF") +
coord_flip() +
theme(legend.position="right") +
scale_x_discrete(labels = wrap_format(40))+
scale_y_continuous(limits= c(1998, 2019), breaks = seq(1998,2020,2)) +
labs(title = "Number of observations for selected SOEP variables from 1998 - 2019",
subtitle = "Size indicates number of observations",
y = "", x = "") 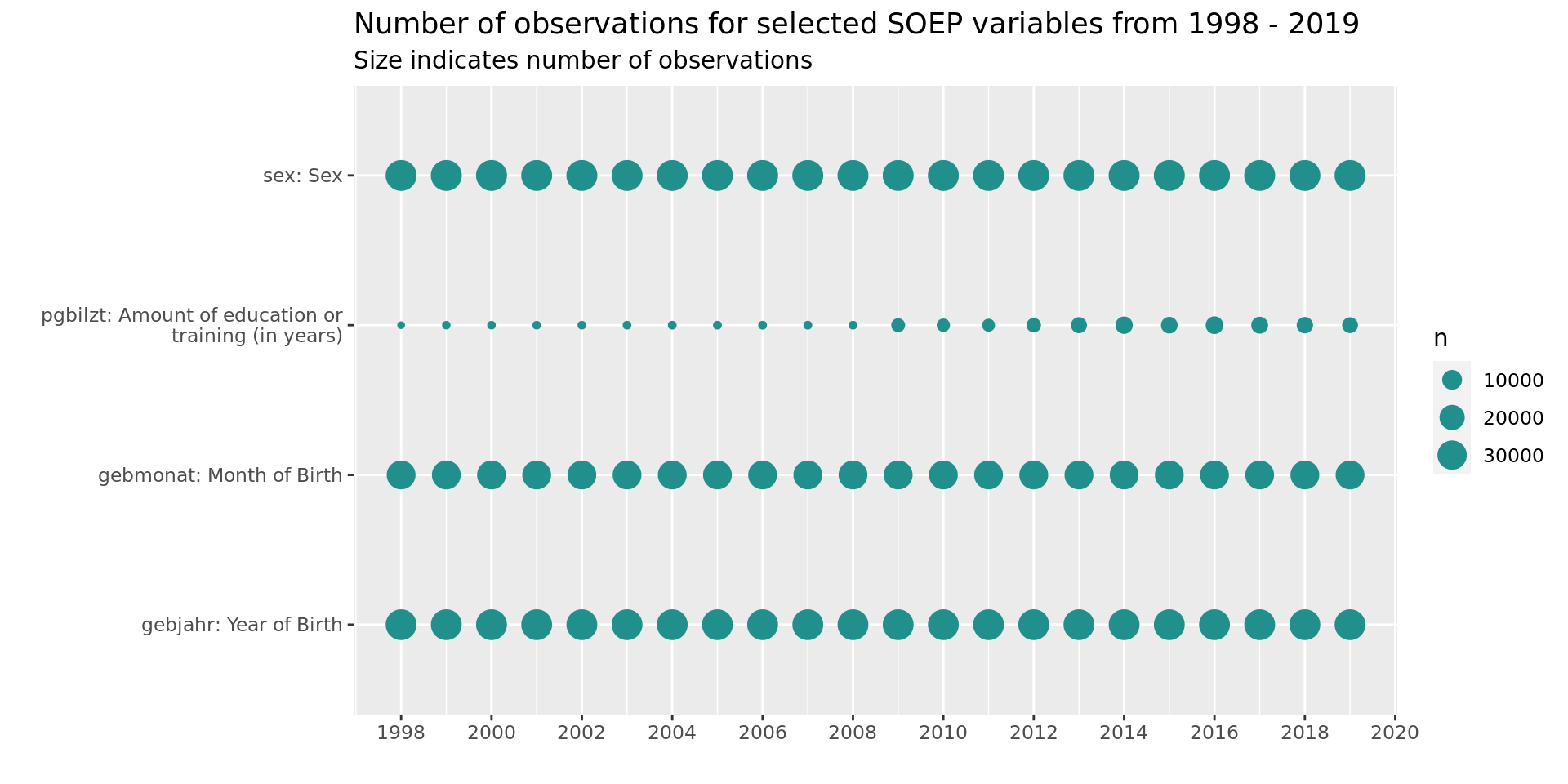
Psychol. Measures
soepis_long %>%
drop_na(value) %>%
filter(key_category == "Psych. Measure") %>%
ggplot(aes(key_name_label, syear)) +
geom_count(col = "#5DC863FF") +
coord_flip() +
theme(legend.position="right") +
scale_x_discrete(labels = wrap_format(40))+
scale_y_continuous(limits= c(1998, 2019), breaks = seq(1998,2020,2)) +
labs(title = "Number of observations for selected SOEP variables from 1998 - 2019",
subtitle = "Size indicates number of observations",
y = "", x = "") 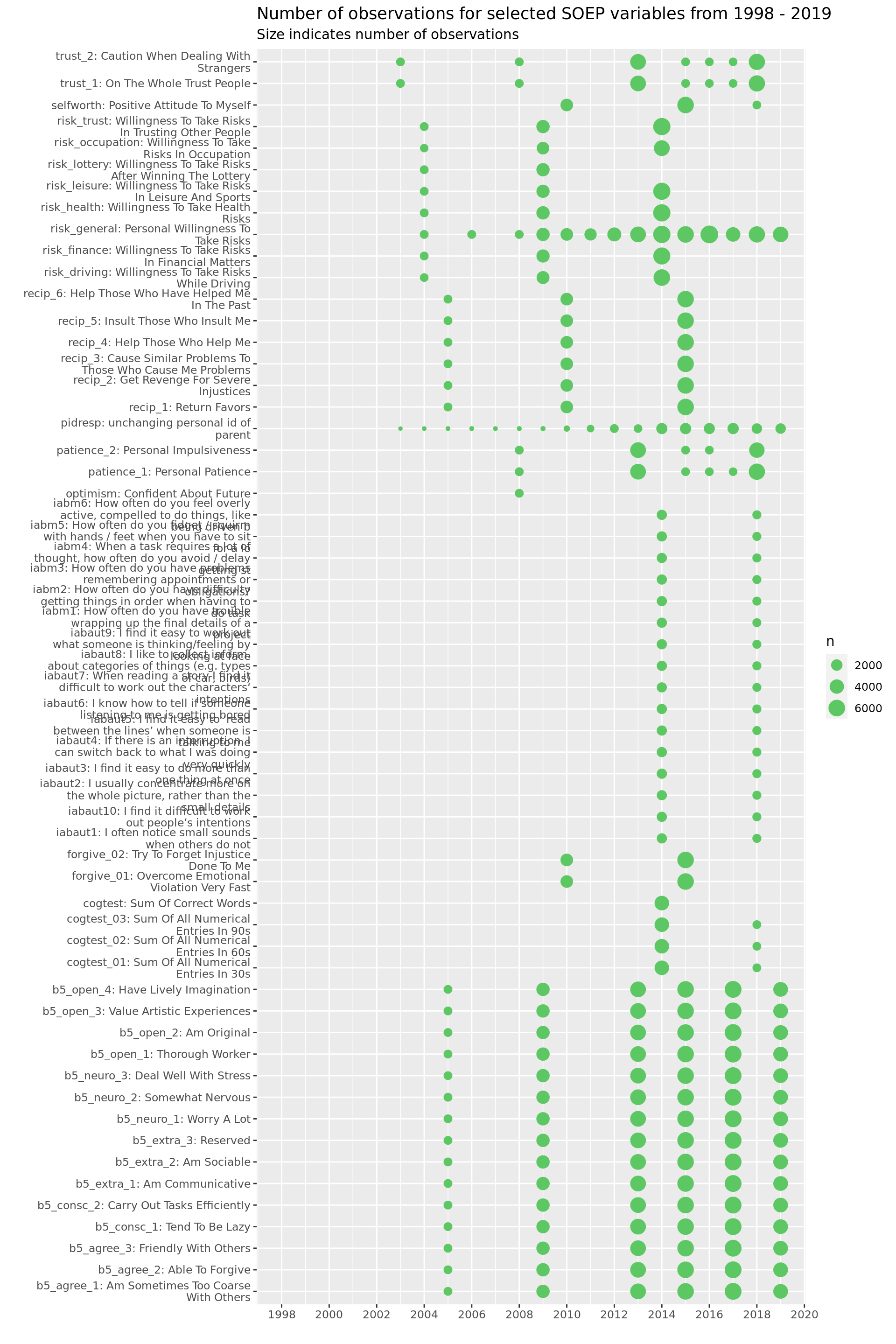
Other
soepis_long %>%
drop_na(value) %>%
filter(key_category == "Other") %>%
ggplot(aes(key_name_label, syear)) +
geom_count(col = "#FDE725FF") +
coord_flip() +
theme(legend.position="right") +
scale_x_discrete(labels = wrap_format(40))+
scale_y_continuous(limits= c(1998, 2019), breaks = seq(1998,2020,2)) +
labs(title = "Number of observations for selected SOEP variables from 1998 - 2019",
subtitle = "Size indicates number of observations",
y = "", x = "") 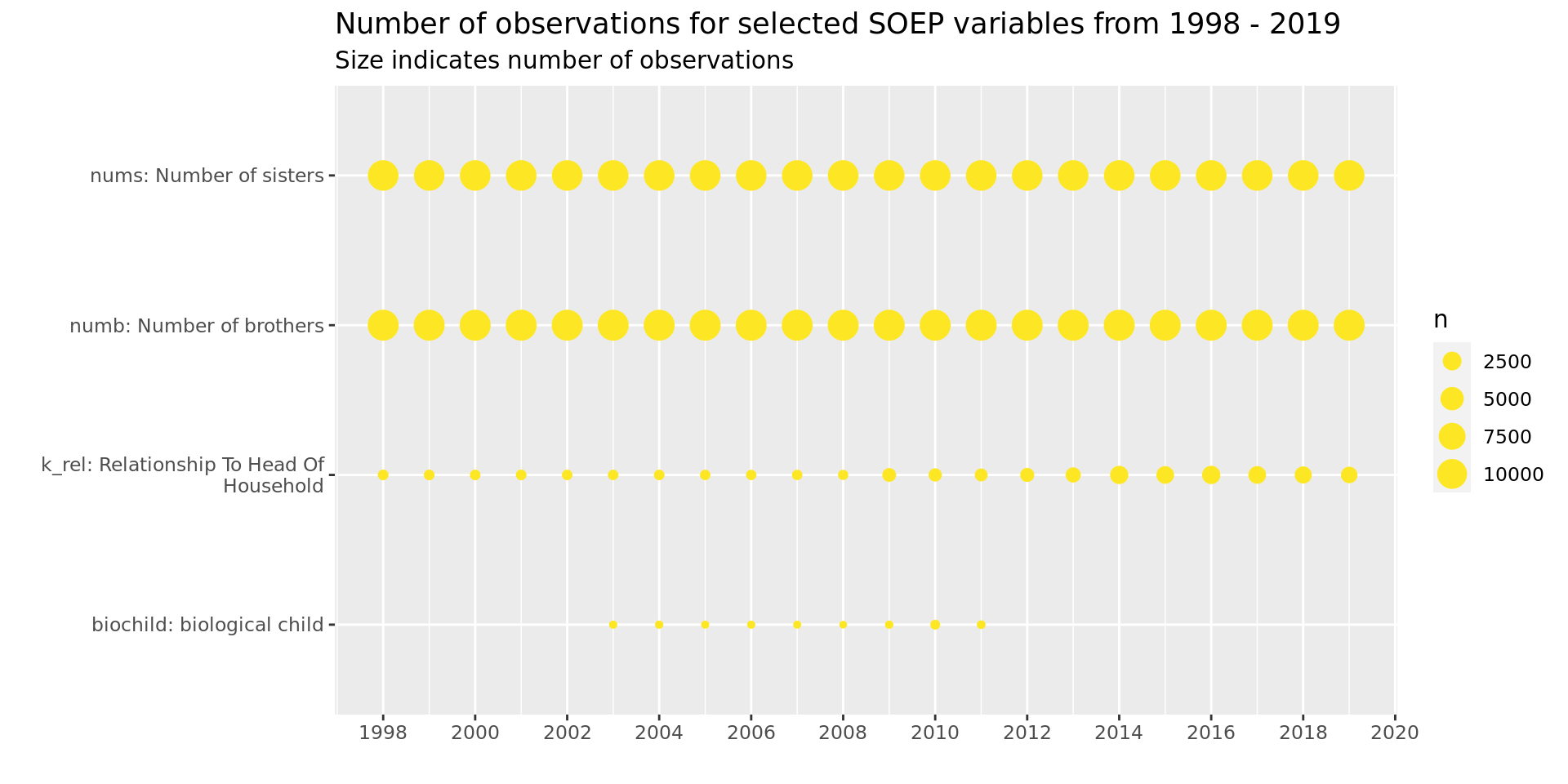
By Age
ID’s
soepis_age %>%
drop_na(value) %>%
filter(age > 0) %>%
# filter(row_number() < 1000) %>%
mutate(age_k = cut_width(age, 15, boundary = 0)) %>%
filter(key_category == "ID's") %>%
ggplot(aes(key_name_label, age)) +
geom_count(col= "#440154FF") +
# scale_size(breaks = seq(0, max(n), 10000)) +
coord_flip() +
theme(legend.position="right") +
scale_x_discrete(labels = wrap_format(40))+
# scale_y_continuous(limits= c(0, 110), breaks = seq(0,110,10))+
labs(title = "Number of observations for selected SOEP variables by age",
subtitle = "Size indicates number of observations",
y = "", x = "") +
facet_wrap(~ age_k, scales = "free_x", nrow = 1, drop = T)+
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank())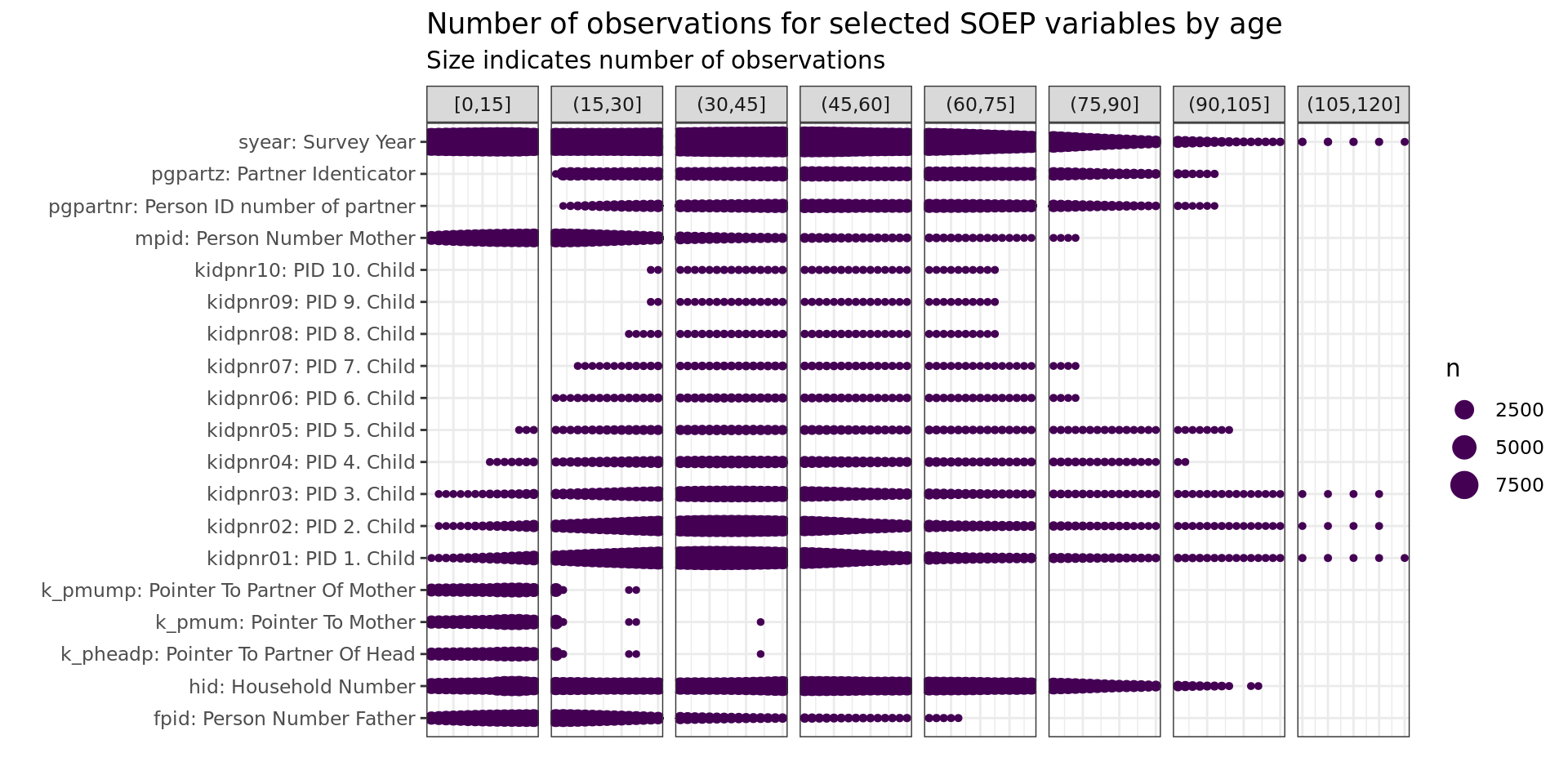
Survey
soepis_age %>%
drop_na(value) %>%
filter(age > 0) %>%
# filter(row_number() < 1000) %>%
mutate(age_k = cut_width(age, 15, boundary = 0)) %>%
filter(key_category == "Survey") %>%
ggplot(aes(key_name_label, age)) +
geom_count(col= "#3B528BFF") +
# scale_size(breaks = seq(0, max(n), 10000)) +
coord_flip() +
theme(legend.position="right") +
scale_x_discrete(labels = wrap_format(40))+
# scale_y_continuous(limits= c(0, 110), breaks = seq(0,110,10))+
labs(title = "Number of observations for selected SOEP variables by age",
subtitle = "Size indicates number of observations",
y = "", x = "") +
facet_wrap(~ age_k, scales = "free_x", nrow = 1, drop = T) +
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank())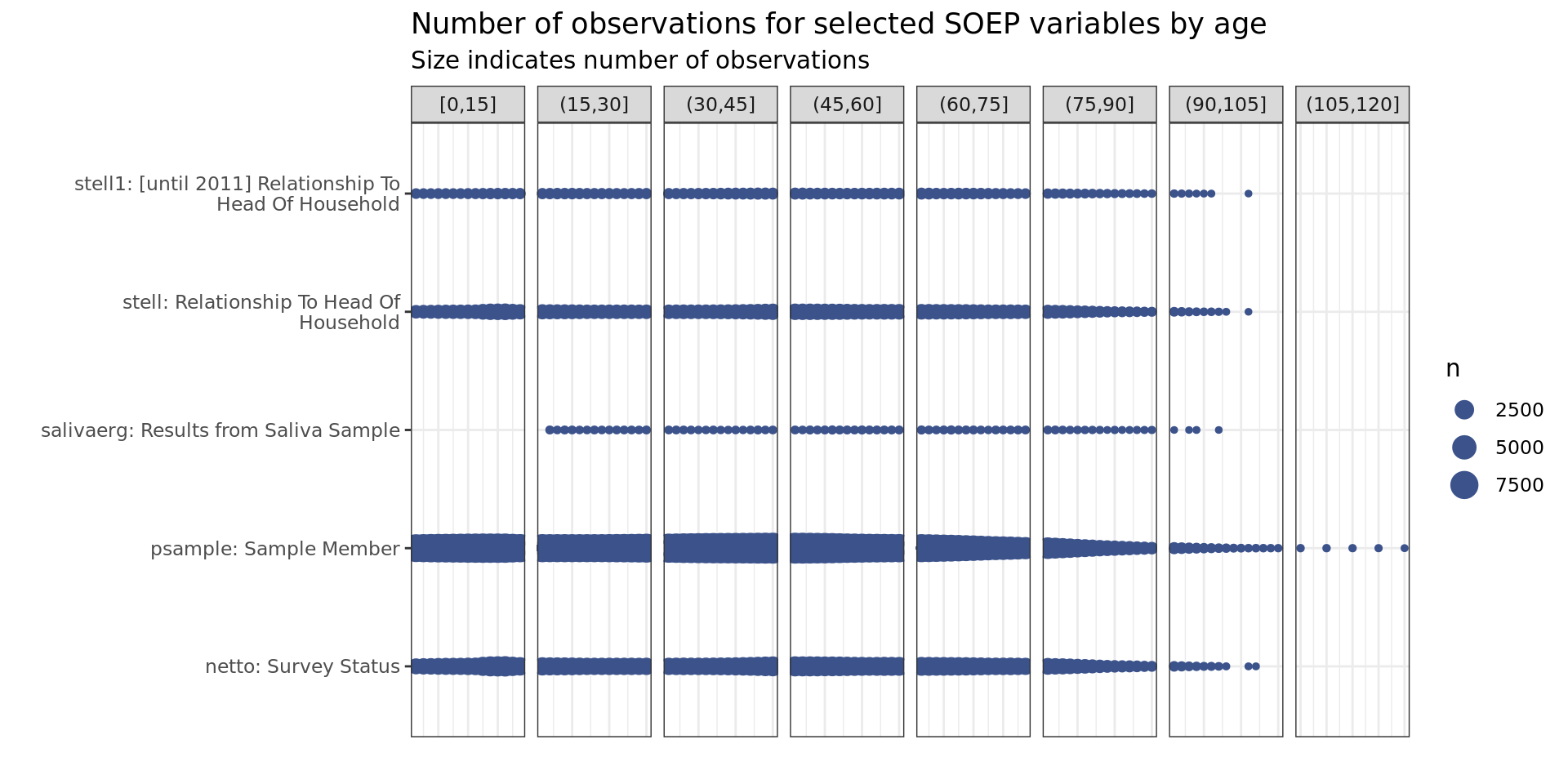
Demography
soepis_age %>%
drop_na(value) %>%
filter(age > 0) %>%
# filter(row_number() < 1000) %>%
mutate(age_k = cut_width(age, 15, boundary = 0)) %>%
filter(key_category == "Demography") %>%
ggplot(aes(key_name_label, age)) +
geom_count(col= "#21908CFF") +
# scale_size(breaks = seq(0, max(n), 10000)) +
coord_flip() +
theme(legend.position="right") +
scale_x_discrete(labels = wrap_format(40))+
# scale_y_continuous(limits= c(0, 110), breaks = seq(0,110,10))+
labs(title = "Number of observations for selected SOEP variables by age",
subtitle = "Size indicates number of observations",
y = "", x = "") +
facet_wrap(~ age_k, scales = "free_x", nrow = 1, drop = T)+
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank())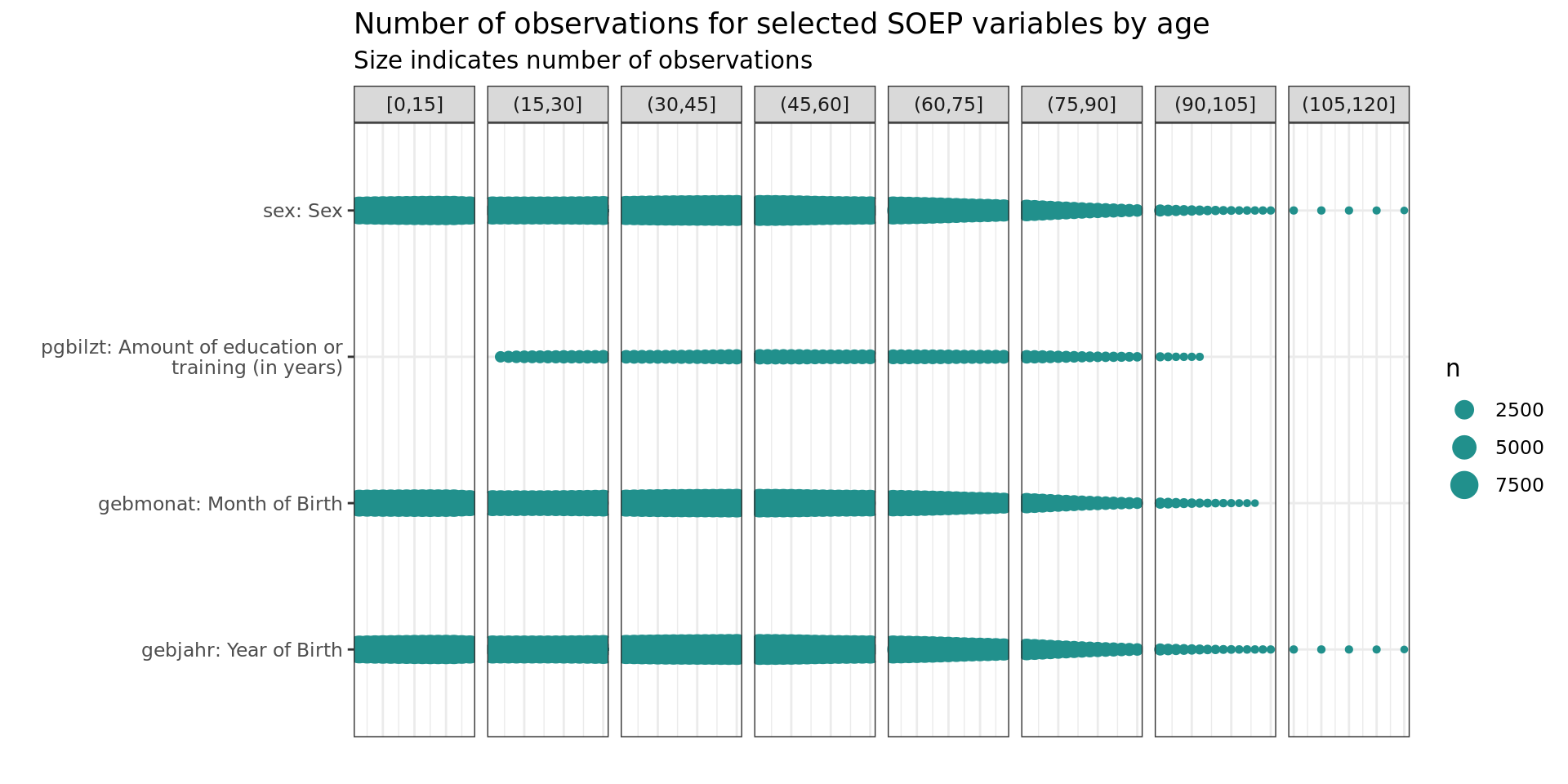
Psychol. Measures
soepis_age %>%
drop_na(value) %>%
filter(age > 0) %>%
# filter(row_number() < 1000) %>%
mutate(age_k = cut_width(age, 15, boundary = 0)) %>%
filter(key_category == "Psych. Measure") %>%
ggplot(aes(key_name_label, age)) +
geom_count(col= "#5DC863FF") +
# scale_size(breaks = seq(0, max(n), 10000)) +
coord_flip() +
theme(legend.position="right") +
scale_x_discrete(labels = wrap_format(40))+
# scale_y_continuous(limits= c(0, 110), breaks = seq(0,110,10))+
labs(title = "Number of observations for selected SOEP variables by age",
subtitle = "Size indicates number of observations",
y = "", x = "") +
facet_wrap(~ age_k, scales = "free_x", nrow = 1, drop = T)+
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank())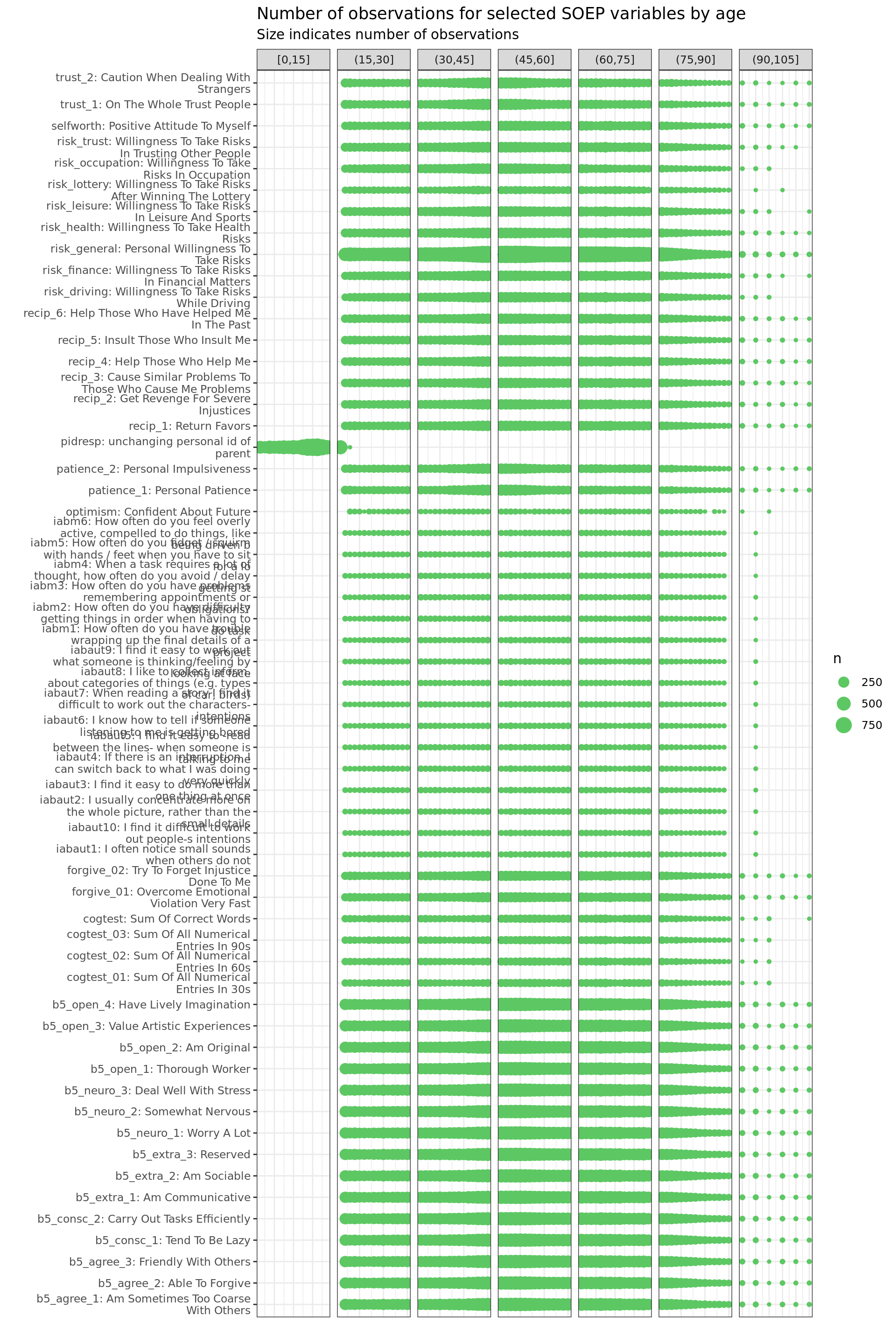
Other
soepis_age %>%
drop_na(value) %>%
filter(age > 0) %>%
# filter(row_number() < 1000) %>%
mutate(age_k = cut_width(age, 15, boundary = 0)) %>%
filter(key_category == "Other") %>%
ggplot(aes(key_name_label, age)) +
geom_count(col= "#FDE725FF") +
# scale_size(breaks = seq(0, max(n), 10000)) +
coord_flip() +
theme(legend.position="right") +
scale_x_discrete(labels = wrap_format(40))+
# scale_y_continuous(limits= c(0, 110), breaks = seq(0,110,10))+
labs(title = "Number of observations for selected SOEP variables by age",
subtitle = "Size indicates number of observations",
y = "", x = "") +
facet_wrap(~ age_k, scales = "free_x", nrow = 1, drop = T)+
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank())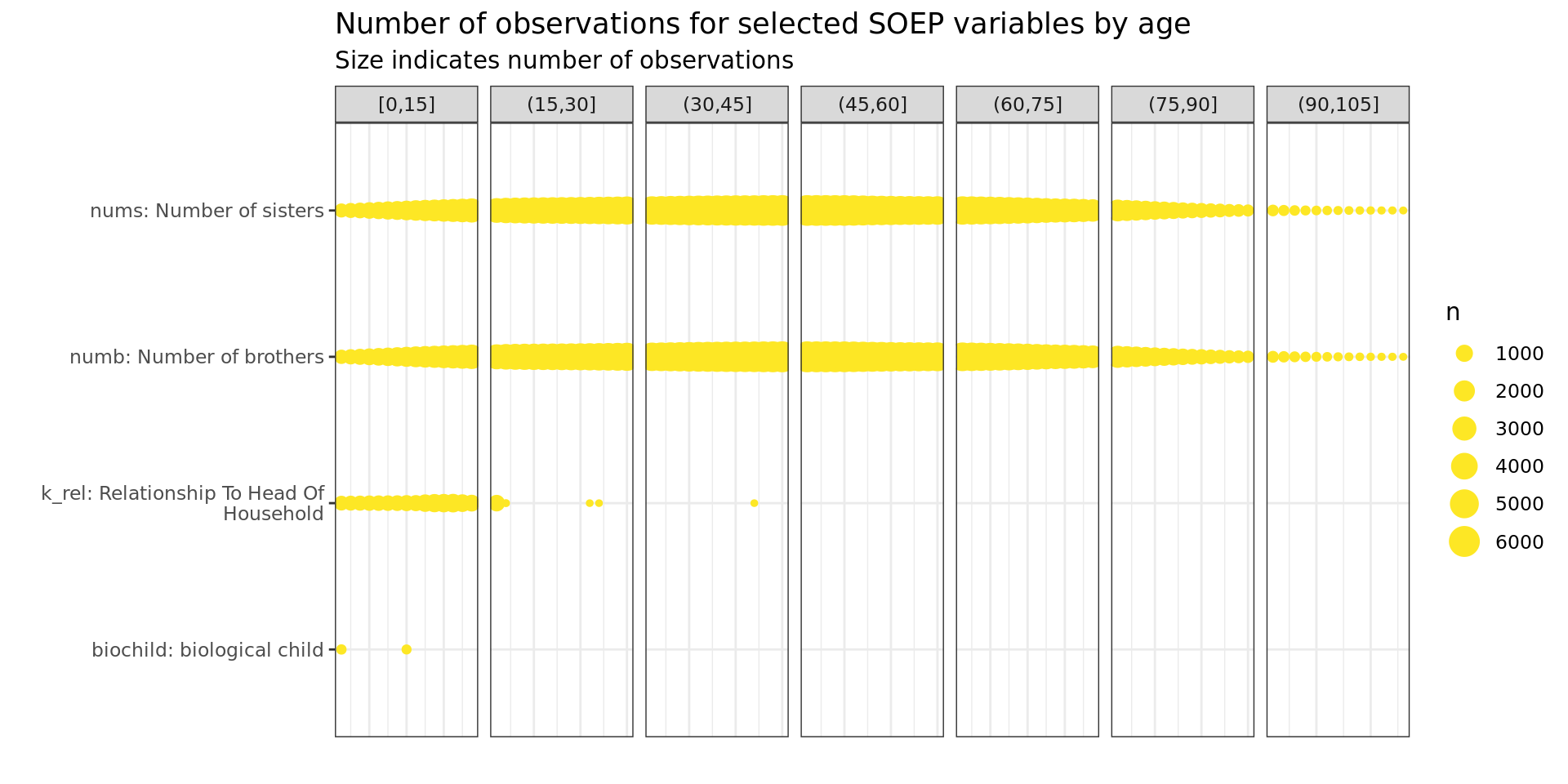
Supplement
resources
rownames to column: https://stackoverflow.com/questions/29511215/convert-row-names-into-first-column age categories: https://ggplot2.tidyverse.org/reference/cut_interval.html wrap label names: https://stackoverflow.com/questions/21878974/auto-wrapping-of-labels-via-labeller-label-wrap-in-ggplot2